5.4. Diffusion with Adsorption in Polymers#
5.4.1. References#
5.4.2. Model#
Here we consider the diffusion of a small molecule into an immobile substrate with adsorption. Following the cited references:
Langmuir isotherm:
After application of the chain rule:
Initial conditions for \(c(t, x)\):
Boundary conditions for \(c(t, x)\):
Exercise 1: Verify the use of the chain rule. Generalize to an arbitrary isotherm \(c = f(c_a)\).
Exercise 2: Compare the dimensional and dimensionless implementations by finding values for dimensionless value of \(\alpha\) and surface concentration that reproduce the simulation results observed in dimensional model.
Exercise 3: Implement a Pyomo model as a DAE without using the chain rule.
Exercise 4: Replace the finite difference transformation on \(x\) with collocation. What (if any) practical advantages are associated with collocation?
Exercise 5: Revise the problem and model with a linear isotherm \(c_a = K c\), and use the solution concentration \(C_s\) to scale \(c(t, x)\). How many independent physical parameters exist for this problem? Find the analytical solution, and compare to a numerical solution found using Pyomo.DAE.
Exercise 6: Revise the dimensionless model for the Langmuir isotherm to use the solution concentraion \(C_s\) to scale \(c(x, t)\). How do the models compare? How many independent parameters exist for this problem? What physical or chemical interpretations can you assign to the dimensionless parameters appearing in the models?
Exercise 7: The Langmuir isotherm is an equilibrium relationship between absorption and desorption processes. The Langmuir isotherm assumes these processes are much faster than diffusion. Formulate the model for the case where absorption and desorption kinetics are on a time scale similar to the diffusion process.
5.4.3. Installations and Imports#
import matplotlib.pyplot as plt
import numpy as np
import shutil
import sys
import os.path
if not shutil.which("pyomo"):
!pip install -q pyomo
assert(shutil.which("pyomo"))
if not (shutil.which("ipopt") or os.path.isfile("ipopt")):
if "google.colab" in sys.modules:
!wget -N -q "https://ampl.com/dl/open/ipopt/ipopt-linux64.zip"
!unzip -o -q ipopt-linux64
else:
try:
!conda install -c conda-forge ipopt
except:
pass
assert(shutil.which("ipopt") or os.path.isfile("ipopt"))
5.4.4. Pyomo model#
import pyomo.environ as pyo
import pyomo.dae as dae
# parameters
tf = 80
D = 2.68
L = 1.0
KL = 20000.0
Cs = 0.0025
qm = 1.0
m = pyo.ConcreteModel()
m.t = dae.ContinuousSet(bounds=(0, tf))
m.x = dae.ContinuousSet(bounds=(0, L))
m.c = pyo.Var(m.t, m.x)
m.dcdt = dae.DerivativeVar(m.c, wrt=m.t)
m.dcdx = dae.DerivativeVar(m.c, wrt=m.x)
m.d2cdx2 = dae.DerivativeVar(m.c, wrt=(m.x, m.x))
@m.Constraint(m.t, m.x)
def pde(m, t, x):
return m.dcdt[t, x] * (1 + qm*KL/(1 + KL*m.c[t, x])** 2) == D * m.d2cdx2[t, x]
@m.Constraint(m.t)
def bc1(m, t):
return m.c[t, 0] == Cs
@m.Constraint(m.t)
def bc2(m, t):
return m.dcdx[t, L] == 0
@m.Constraint(m.x)
def ic(m, x):
if x == 0:
return pyo.Constraint.Skip
return m.c[0, x] == 0.0
# transform and solve
pyo.TransformationFactory('dae.finite_difference').apply_to(m, wrt=m.x, nfe=40)
pyo.TransformationFactory('dae.finite_difference').apply_to(m, wrt=m.t, nfe=40)
pyo.SolverFactory('ipopt').solve(m).write()
# ==========================================================
# = Solver Results =
# ==========================================================
# ----------------------------------------------------------
# Problem Information
# ----------------------------------------------------------
Problem:
- Lower bound: -inf
Upper bound: inf
Number of objectives: 1
Number of constraints: 6682
Number of variables: 6683
Sense: unknown
# ----------------------------------------------------------
# Solver Information
# ----------------------------------------------------------
Solver:
- Status: ok
Message: Ipopt 3.13.4\x3a Optimal Solution Found
Termination condition: optimal
Id: 0
Error rc: 0
Time: 1.7579870223999023
# ----------------------------------------------------------
# Solution Information
# ----------------------------------------------------------
Solution:
- number of solutions: 0
number of solutions displayed: 0
5.4.5. Visualization#
def model_plot(m):
t = sorted(m.t)
x = sorted(m.x)
xgrid = np.zeros((len(t), len(x)))
tgrid = np.zeros((len(t), len(x)))
cgrid = np.zeros((len(t), len(x)))
for i in range(0, len(t)):
for j in range(0, len(x)):
xgrid[i,j] = x[j]
tgrid[i,j] = t[i]
cgrid[i,j] = m.c[t[i], x[j]].value
fig = plt.figure(figsize=(10,6))
ax = fig.add_subplot(1, 1, 1, projection='3d')
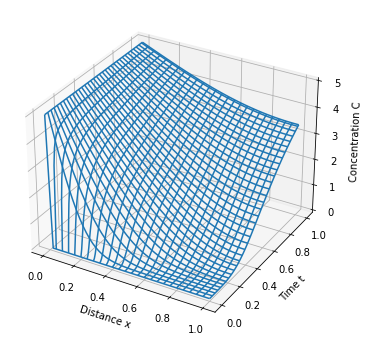
ax.set_xlabel('Distance x')
ax.set_ylabel('Time t')
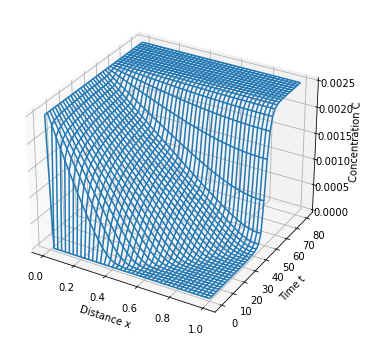
ax.set_zlabel('Concentration C')
p = ax.plot_wireframe(xgrid, tgrid, cgrid)
# visualization
model_plot(m)
5.4.6. Dimensional analysis#
Assuming \(L\) is determined by the experimental apparatus, choose
which results in a one parameter model
where \(\alpha = q K\) represents a dimensionless capacity of the substrate to absorb the diffusing molecule.
5.4.7. Dimensionless Pyomo model#
import pyomo.environ as pyo
import pyomo.dae as dae
# parameters
tf = 1.00
Cs = 5.0
alpha = 5.0
m = pyo.ConcreteModel()
m.t = dae.ContinuousSet(bounds=(0, tf))
m.x = dae.ContinuousSet(bounds=(0, 1))
m.c = pyo.Var(m.t, m.x)
m.s = pyo.Var(m.t, m.x)
m.dcdt = dae.DerivativeVar(m.c, wrt=m.t)
m.dcdx = dae.DerivativeVar(m.c, wrt=m.x)
m.d2cdx2 = dae.DerivativeVar(m.c, wrt=(m.x, m.x))
@m.Constraint(m.t, m.x)
def pde(m, t, x):
return m.dcdt[t, x] * (1 + alpha/(1 + m.c[t, x])** 2) == m.d2cdx2[t, x]
@m.Constraint(m.t)
def bc1(m, t):
return m.c[t, 0] == Cs
@m.Constraint(m.t)
def bc2(m, t):
return m.dcdx[t, 1] == 0
@m.Constraint(m.x)
def ic(m, x):
if (x == 0) or (x == 1):
return pyo.Constraint.Skip
return m.c[0, x] == 0.0
# transform and solve
pyo.TransformationFactory('dae.finite_difference').apply_to(m, wrt=m.x, nfe=50)
pyo.TransformationFactory('dae.finite_difference').apply_to(m, wrt=m.t, nfe=100)
pyo.SolverFactory('ipopt').solve(m).write()
model_plot(m)
# ==========================================================
# = Solver Results =
# ==========================================================
# ----------------------------------------------------------
# Problem Information
# ----------------------------------------------------------
Problem:
- Lower bound: -inf
Upper bound: inf
Number of objectives: 1
Number of constraints: 20501
Number of variables: 20503
Sense: unknown
# ----------------------------------------------------------
# Solver Information
# ----------------------------------------------------------
Solver:
- Status: ok
Message: Ipopt 3.13.4\x3a Optimal Solution Found
Termination condition: optimal
Id: 0
Error rc: 0
Time: 3.6713459491729736
# ----------------------------------------------------------
# Solution Information
# ----------------------------------------------------------
Solution:
- number of solutions: 0
number of solutions displayed: 0