RPI Library tests
Contents
RPI Library tests#
Show numpy configuration#
import numpy as np
# np.show_config()
Matrix Multiplication Benchmark#
import numpy as np
from time import time
np.random.seed(0)
size = 1024
A, B = np.random.random((size, size)), np.random.random((size, size))
# Matrix multiplication
N = 10
t = time()
for i in range(N):
np.dot(A, B)
delta = time() - t
print('Dotted two %dx%d matrices in %0.2f s.' % (size, size, delta / N))
del A, B
Dotted two 1024x1024 matrices in 0.31 s.
Matplotlib Demonstration#
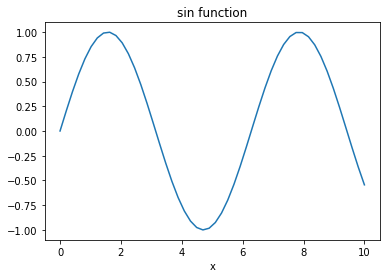
import matplotlib.pyplot as plt
x = np.linspace(0, 10)
fig, ax = plt.subplots(1, 1)
ax.plot(x, np.sin(x))
ax.set_xlabel('x')
ax.set_title('sin function')
Text(0.5, 1.0, 'sin function')
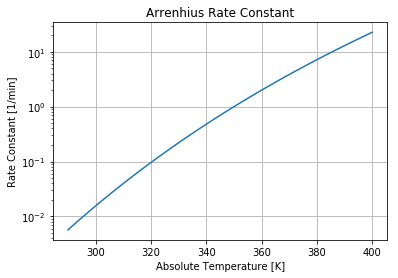
Scipy Demonstration#
import matplotlib.pyplot as plt
import numpy as np
# Arrehnius parameters
Ea = 72750 # activation energy J/gmol
R = 8.314 # gas constant J/gmol/K
k0 = 7.2e10 # Arrhenius rate constant 1/min
# Arrhenius rate expression
def k(T):
return k0*np.exp(-Ea/(R*T))
# semilog plot of the rate constant
T = np.linspace(290,400)
plt.semilogy(T, k(T))
plt.xlabel('Absolute Temperature [K]')
plt.ylabel('Rate Constant [1/min]')
plt.title('Arrenhius Rate Constant')
plt.grid();
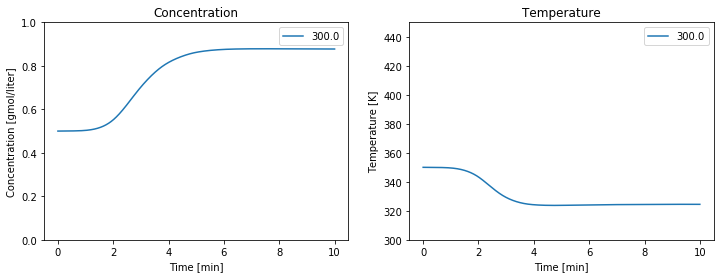
from scipy.integrate import solve_ivp
Ea = 72750 # activation energy J/gmol
R = 8.314 # gas constant J/gmol/K
k0 = 7.2e10 # Arrhenius rate constant 1/min
V = 100.0 # Volume [L]
rho = 1000.0 # Density [g/L]
Cp = 0.239 # Heat capacity [J/g/K]
dHr = -5.0e4 # Enthalpy of reaction [J/mol]
UA = 5.0e4 # Heat transfer [J/min/K]
q = 100.0 # Flowrate [L/min]
cAi = 1.0 # Inlet feed concentration [mol/L]
Ti = 350.0 # Inlet feed temperature [K]
cA0 = 0.5; # Initial concentration [mol/L]
T0 = 350.0; # Initial temperature [K]
Tc = 300.0 # Coolant temperature [K]
def deriv(t, y):
cA,T = y
dcAdt = (q/V)*(cAi - cA) - k(T)*cA
dTdt = (q/V)*(Ti - T) + (-dHr/rho/Cp)*k(T)*cA + (UA/V/rho/Cp)*(Tc-T)
return [dcAdt, dTdt]
# simulation
IC = [cA0, T0]
t_initial = 0.0
t_final = 10.0
t = np.linspace(t_initial, t_final, 2000)
soln = solve_ivp(deriv, [t_initial, t_final], IC, t_eval=t)
# visualization plots concentration and temperature on given axes
def plot_reactor(ax, t, y):
ax[0].plot(t, y[0], label=str(Tc))
ax[0].set_xlabel('Time [min]')
ax[0].set_ylabel('Concentration [gmol/liter]')
ax[0].set_title('Concentration')
ax[0].set_ylim(0, 1)
ax[0].legend()
ax[1].plot(t, y[1], label=str(Tc))
ax[1].set_xlabel('Time [min]')
ax[1].set_ylabel('Temperature [K]');
ax[1].set_title('Temperature')
ax[1].set_ylim(300, 450)
ax[1].legend()
fig, ax = plt.subplots(1, 2, figsize=(12, 4))
plot_reactor(ax, soln.t, soln.y);
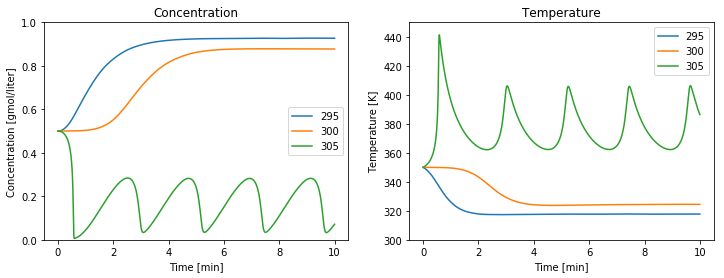
fig, ax = plt.subplots(1, 2, figsize=(12, 4))
for Tc in [295, 300, 305]:
soln = solve_ivp(deriv, [t_initial, t_final], IC, t_eval=t)
plot_reactor(ax, soln.t, soln.y)